The ribosomes are the most abundant protein complexes in the cell. They are synthesised from ribosomal DNA (rDNA) genes within nucleoli. In most eukaryotic cells, rDNA exists in tandem arrays, which are repeating units of similar sequences. Oftentimes the rDNA provide sites for recombination, and the copy number seems to fluctuate. In this regard, the rDNA is hypothesised to be involved in genome stability and cellular senescence.
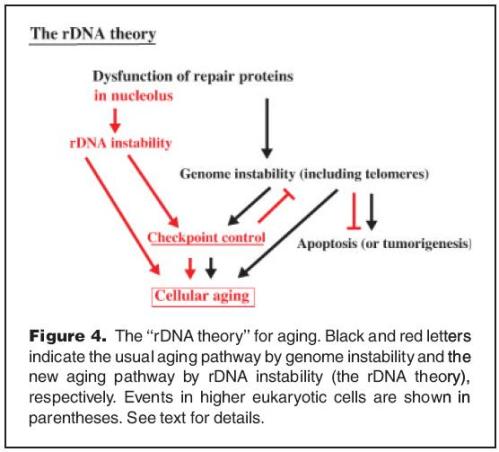
There was a paper written by Takehiko Kobayashi last year, 2008, in the journal BioEssays, wherein he proposed the rDNA theory of aging, stating that the rDNA may be the region most sensitive to DNA damage. He highlighted two key molecular entities that would link rDNA to aging: FOB1 and SIR2. These two are well known aging genes.
“… in yeast Saccharomyces cerevisiae … lifespan expansion in a fob1 mutant is associated with increased rDNA stability (no copy number variation), while the lifespan reduction in a sir2 mutant is associated with decreased rDNA stability (high copy number variation) …”
“rDNA is more unstable and induces checkpoint control faster than any other part of genome, and that the nucleolus may be more sensitive to protein damage because it is protein-dense …”
He stated further that rDNA may serve as ‘‘sensor’’ for DNA damage, and also as ‘‘shock absorber’’ that protects the genome from damage. I think these are broad statements as of this time.
He and his colleagues have observed that fob1 mutants show better growth than wild type, indicating checkpoint induction is reduced and lifespan is extended. More specifically, he mentioned that rDNA copy number gradually decreases in a fob1 mutant by a FOB1-independent form of homologous recombination. This implies termination of growth, presumbaly because of a shortage of rRNA and ribosomes. In this manner, lifespan extends due growth delay or termination. And yet, the number of abnormal cells is likely to increase.
In contrast, he described that sir2 mutants show increased rDNA instability. He stated that this leads to greater checkpoint control and shorter lifespan. I shall look further into this as it seems unclear to me.
Below is one of the images he presented in the paper:
A new role of the rDNA and nucleolus in the nucleus – rDNA instability maintains genome integrity
Takehiko Kobayashi
National Institute of Genetics and The Graduate University for Advanced Studies, SOKENDAI, 1111 Yata, Mishima 411-8540 Japan
email: Takehiko Kobayashi (takobaya@lab.nig.ac.jp)
BioEssays (2008)
Volume 30 Issue 3, Pages 267 – 272